PCR is an enzymatic amplification process which requires DNA or RNA polymerase, primers and deoxynucleotide triphosphates in an appropriate buffer, and means of controlling the temperature during the various stages of the amplification process. The primers are short single-stranded oligonucleotide sequences complimentary to the 3′ ends flanking the segment of DNA to be amplified. During PCR, the template DNA is denatured to single-strands at a high temperature, then allowed to anneal with an excess of primers at a lower temperature, followed by synthesis of DNA or RNA from the primers at a temperature optimum for the specific polymerase enzyme. After synthesis, the cycle of denaturation, annealing and synthesis is repeated whereby the products of the previous amplification reaction serve as templates for the next round of amplification. This cycle is repeated for as many as 30 times leading to the exponential amplification of the DNA segment of interest. The major product of the PCR amplification process is a double-stranded segment of DNA whose length is defined by the distance between the two primers used and whose termini correspond to the 5′ termini of the primers. The changes in temperature during the cycles can be achieved by using commercially available automated thermal cyclers or by using a series of temperature-controlled water baths. Automated thermocyclers are now in wide in use as they are convenient.
Isolation of DNA from cheek cells:
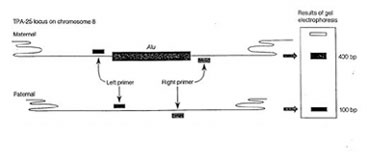
The source of template DNA for amplification is a sample of several thousand cells obtained by saline mouthwash (bloodless and noninvasive). The cells are collected by centrifugation and resuspended in a solution containing the resin “Chelex. The suspension (containing Chelex) is then boiled to lyse the cells. Following cell lysis Chelex will bind to and remove the proteins and metal ions that inhibit the PCR reaction, but will leave the DNA in the solution. The cell debris and the resin beads are removed by centrifugation. A sample of the supernatant containing chromosomal DNA is mixed with Taq DNA polymerase, oligonucleotide primers, the four deoxyribonucleotides, and the cofactor magnesium chloride. Temperature cycling is used to denature the target DNA, anneal the primers, and extend a complementary DNA strand. The size of the amplification product(s) depends on the presence or absence of the Alu insertion at the TPA-25 locus on each copy of chromosome 8.
Isolate Cheek Cell DNA for PCR Amplification:
1. Add 6 ml of saline solution to a 10 ml screw capped tube.
2. Pour all of the saline solution into your mouth and vigorously swish for 10 seconds. Do not discard the empty tube that contained the saline solution.
3. Expel the sample solution into a paper cup.
4. Pour the sample solution from the paper cup back into the tube that contained the saline solution and close cap tightly.
5. Spin sample in preparatory centrifuge on high speed for 10 minutes.
6. Carefully pour off supernatant (liquid on top) into sink and place tube containing your cells on ice.
7. Set the P1000 micropipettor to 500 µl and pipet the Chelex solution in and out of the pipet tip several times to suspend the Chelex beads. Before the Chelex has a chance to settle, add the 500 µl to the centrifuge tube containing your cell pellet.
8. Mix cells and Chelex by pipeting up and down several times until no visible clumps of cells remain.
9. Using the same pipet tip, transfer 500 µl of your resuspended sample into a clean 1.5 ml tube. Be sure to label the cap of the tube with your initials.
10. Place your tube in a boiling water bath for ten minutes.
12. Carefully remove your tube from the boiling water bath and place on ice for one minute.
13. Place your tube in a microcentrifuge (opposite someone else1s tube!) and centrifuge for 30 seconds.
14. Use a fresh pipet tip to transfer 200 µl of supernatant (clear solution on top) to another clean 1.5 ml tube. Label with your name.
15. This contain your DNA which you can use as the template for amplification by the polymerase chain reaction.
Detection of an Alu insertion polymorphism by PCR:
Some regions of the human genome show variation between individuals. Such variations in sequence are called polymorphisms and are made to practical use in diagnosis of genetic diseases , forensic identification and paternity testing. In this experiment you would use DNA isolated from your cheek cells (see above) as template to amplify a short region of DNA within the plasmogen activator gene (TPA) containing a short 300bp insertion called an Alu element. Alu elements are short interspersed, repeated DNA elements distributed through out the primate genomes. The Alu element found within an intron of the TPA gene is dimorphic, i.e. it is present in some individuals but not in others. Following PCR amplification and electrophoresis you would be able to determine if the individual is heterozygous or homozygous for the presence or absence of the Alu insertion.
1. Prepare PCR reaction and load into PCR machine.
2. Mix by tapping the tubes gently.
3. Spin down briefly in the microcentrifuge.
4. Add 100ul of mineral oil on top of the reaction mixtures and place the tubes on ice.
5. Leave tube # A4 at room temperature for 15 min.
6. Place all the tubes in the DNA Thermal Cycler and initiate the program specified for the DNA samples in use.
Gel Electrophoresis of PCR amplified DNA
7. Prepare a 1% agarose minigel in TBE buffer.
8. At the end of the PCR reaction, take a 10 ml aliquot of each reaction without including the oil overlay and transfer to 1.5 ml microtubes.
9. Add 2 ml of gel loading buffer to each 10 ml aliquot. Load in gel along with 1kb ladder molecular weight marker.
10. Run the gel for 1 hour at 80 Volts constant voltage.
11. Stain, destain and photograph the gel as usual.
12. Analyze the DNA fragments on the gel.
